Download this example as Julia file or Jupyter notebook.
SW18 - Distorted kagome
This is a Sunny port of SpinW Tutorial 18, originally authored by Goran Nilsen and Sandor Toth. This tutorial illustrates spin wave calculations for KCu₃As₂O₇(OD)₃. The Cu ions are arranged in a distorted kagome lattice and exhibit an incommensurate helical magnetic order, as described in G. J. Nilsen, et al., Phys. Rev. B 89, 140412 (2014). The model follows Toth and Lake, J. Phys.: Condens. Matter 27, 166002 (2015).
using Sunny, GLMakieBuild the distorted kagome crystal, with spacegroup 12 ("C 1 2/m 1" setting).
units = Units(:meV, :angstrom)
latvecs = lattice_vectors(10.2, 5.94, 7.81, 90, 117.7, 90)
positions = [[0, 0, 0], [1/4, 1/4, 0]]
types = ["Cu1", "Cu2"]
cryst = Crystal(latvecs, positions, "C 1 2/m 1"; types)
view_crystal(cryst)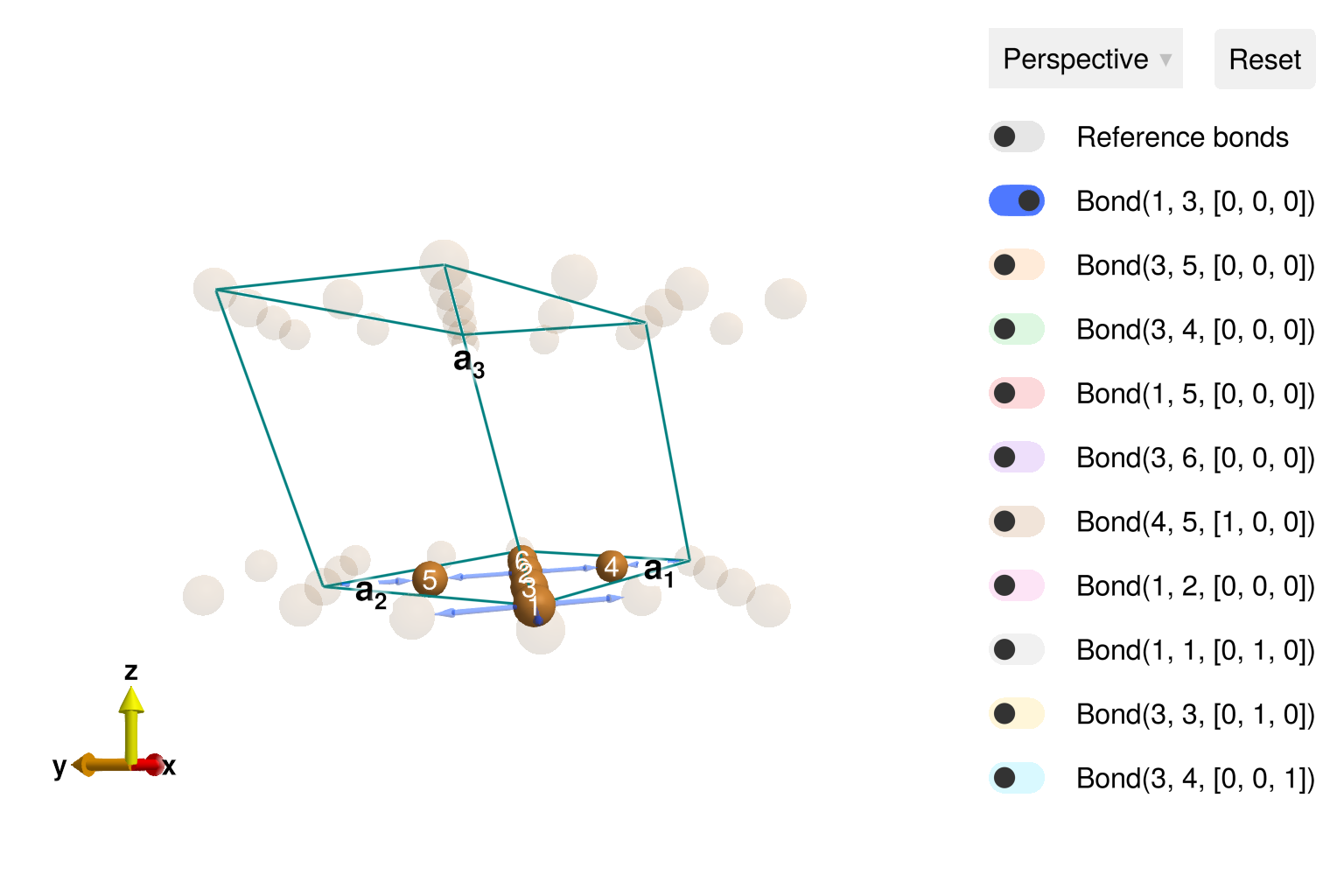
Define the interactions.
moments = [1 => Moment(s=1/2, g=2), 3 => Moment(s=1/2, g=2)]
sys = System(cryst, moments, :dipole)
J = -2
Jp = -1
Jab = 0.75
Ja = -J/.66 - Jab
Jip = 0.01
set_exchange!(sys, J, Bond(1, 3, [0, 0, 0]))
set_exchange!(sys, Jp, Bond(3, 5, [0, 0, 0]))
set_exchange!(sys, Ja, Bond(3, 4, [0, 0, 0]))
set_exchange!(sys, Jab, Bond(1, 2, [0, 0, 0]))
set_exchange!(sys, Jip, Bond(3, 4, [0, 0, 1]))Use minimize_spiral_energy! to optimize the generalized spiral order. This determines the propagation wavevector k and fits the spin values within the unit cell. One must provide a fixed axis perpendicular to the polarization plane. For this system, all interactions are rotationally invariant and the axis vector is arbitrary. In other cases, a good axis will frequently be determined from symmetry considerations.
axis = [0, 0, 1]
randomize_spins!(sys)
k = minimize_spiral_energy!(sys, axis; k_guess=randn(3))
plot_spins(sys; ndims=2)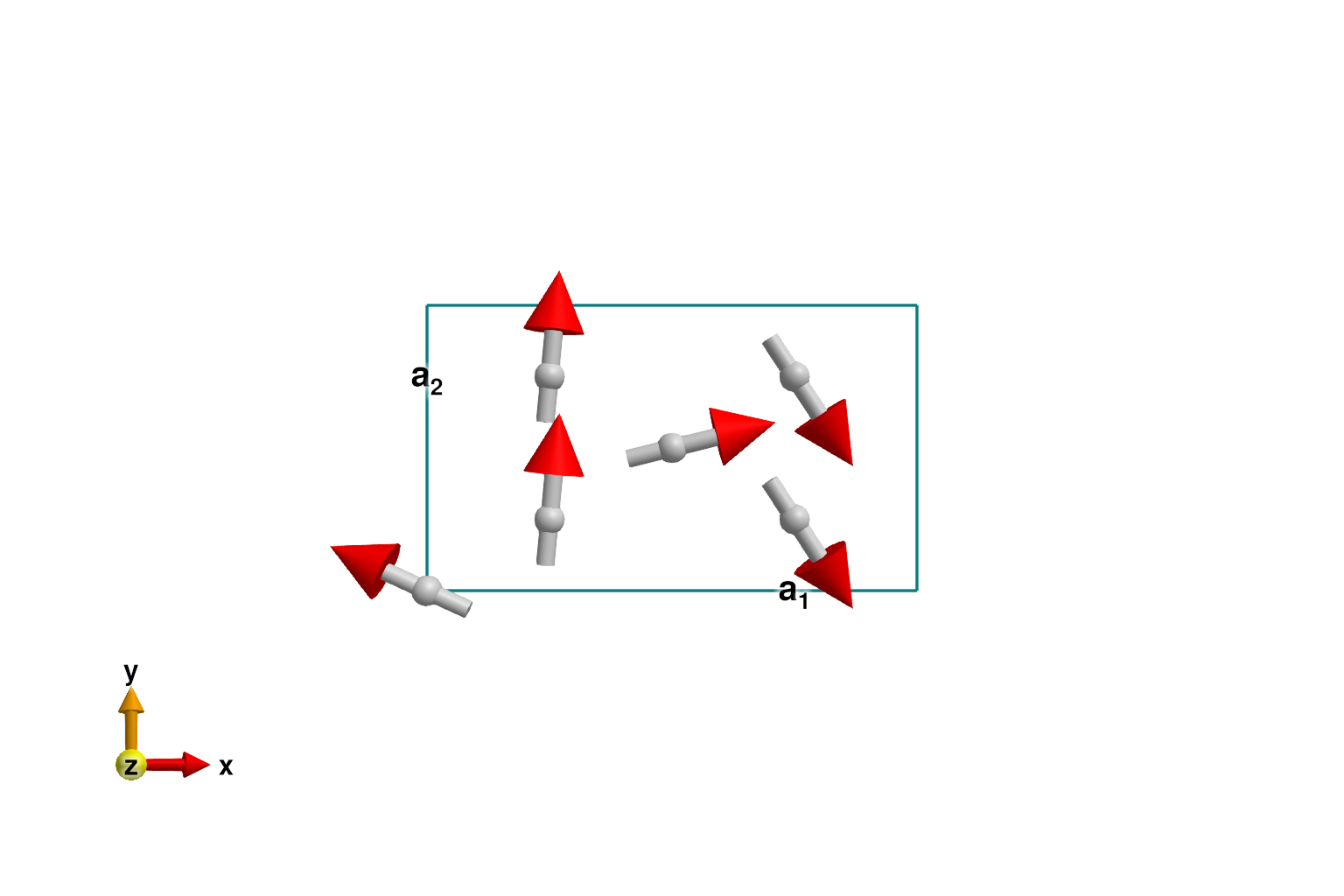
If successful, the optimization process will find one two propagation wavevectors, ±k_ref, with opposite chiralities. In this system, the spiral_energy_per_site is independent of chirality.
k_ref = [0.785902495, 0.0, 0.107048756]
k_ref_alt = [1, 0, 1] - k_ref
@assert isapprox(k, k_ref; atol=1e-6) || isapprox(k, k_ref_alt; atol=1e-6)
@assert spiral_energy_per_site(sys; k, axis) ≈ -0.78338383838Check the energy with a real-space calculation using a large magnetic cell. First, we must determine a lattice size for which k becomes approximately commensurate.
suggest_magnetic_supercell([k_ref]; tol=1e-3)Possible magnetic supercell in multiples of lattice vectors:
[1 0 -7; 0 1 0; 2 0 14]
for the rationalized wavevectors:
[[11/14, 0, 3/28]]Resize the system as suggested and perform a real-space calculation. Working with a commensurate wavevector increases the energy slightly. The precise value might vary from run-to-run due to trapping in a local energy minimum.
new_shape = [14 0 1; 0 1 0; 0 0 2]
sys2 = reshape_supercell(sys, new_shape)
randomize_spins!(sys2)
minimize_energy!(sys2)
energy_per_site(sys2)-0.7833837597216421Return to the original system (with a single chemical cell) and construct SpinWaveTheorySpiral for calculations on the incommensurate spiral phase.
measure = ssf_perp(sys; apply_g=false)
swt = SpinWaveTheorySpiral(sys; measure, k, axis)SpinWaveTheorySpiral(SpinWaveTheory(System([Dipole mode], Supercell (1×1×1)×6, Energy per site -0.3888), Sunny.SWTDataDipole(StaticArraysCore.SMatrix{3, 3, Float64, 9}[[0.21110511458084164 0.3599329917949192 -0.9087809813235357; 0.4597664084417473 0.7838990506971835 0.4172734450988124; 0.8625830280421627 -0.5059155262824159 -1.048990684667789e-15], [0.01010814056795529 -0.2395910448623525 0.9708212794927936; -0.04092171406769344 0.9699584372713026 0.23980417695272813; -0.9991112244450168 -0.0421516451396574 -2.024541475366478e-12], [0.919239023096581 -0.38231339694440214 0.09400045177165757; -0.08679319028227307 0.0360974660315771 0.9955721546260343; -0.384013750453117 -0.923327373937831 2.671257829588084e-12], [0.824002424578344 0.14863927847658134 0.5467415926952803; 0.5380576207846907 0.09705856942326088 -0.8373013978353483; -0.17752183247287817 0.9841168624688185 -1.3899379944907387e-12], [0.824961496858891 -0.5573171841661585 0.0940004517725441; -0.07789164556071708 0.052621065033748735 0.9955721546259506; -0.5597958737362949 -0.8286305447832697 2.5831061214328467e-12], [0.8371073023747876 -0.018027621275770664 0.54674159269434; 0.546614852118133 -0.01177168746676366 -0.8373013978359624; 0.021530623648138465 0.9997681892545502 -2.004557460923225e-12]], StaticArraysCore.SVector{3, Float64}[[0.21110511458084164, 0.3599329917949192, -0.9087809813235357] [0.01010814056795529, -0.2395910448623525, 0.9708212794927936] … [0.824961496858891, -0.5573171841661585, 0.0940004517725441] [0.8371073023747876, -0.018027621275770664, 0.54674159269434]; [0.4597664084417473, 0.7838990506971835, 0.4172734450988124] [-0.04092171406769344, 0.9699584372713026, 0.23980417695272813] … [-0.07789164556071708, 0.052621065033748735, 0.9955721546259506] [0.546614852118133, -0.01177168746676366, -0.8373013978359624]; [0.8625830280421627, -0.5059155262824159, -1.048990684667789e-15] [-0.9991112244450168, -0.0421516451396574, -2.024541475366478e-12] … [-0.5597958737362949, -0.8286305447832697, 2.5831061214328467e-12] [0.021530623648138465, 0.9997681892545502, -2.004557460923225e-12]], Sunny.StevensExpansion[Sunny.StevensExpansion([0.0], [0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], 0), Sunny.StevensExpansion([0.0], [0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], 0), Sunny.StevensExpansion([0.0], [0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], 0), Sunny.StevensExpansion([0.0], [0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], 0), Sunny.StevensExpansion([0.0], [0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], 0), Sunny.StevensExpansion([0.0], [0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], 0)], [0.7071067811865476, 0.7071067811865476, 0.7071067811865476, 0.7071067811865476, 0.7071067811865476, 0.7071067811865476]), MeasureSpec, 1.0e-8), [0.2140975054608815, -2.094959386546427e-13, 0.8929512472833465], 3, [0.0, 0.0, 1.0], Matrix{StaticArraysCore.SMatrix{3, 3, ComplexF64, 9}}[[[0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] … [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im]; [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] … [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im]; … ; [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] … [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im]; [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] … [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im]], [[0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] … [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im]; [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] … [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im]; … ; [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] … [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im]; [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] … [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im]], [[0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] … [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im]; [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] … [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im]; … ; [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] … [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im]; [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] … [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im]], [[0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] … [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im]; [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] … [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im]; … ; [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] … [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im]; [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] … [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im]], [[0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] … [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im]; [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] … [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im]; … ; [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] … [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im]; [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] … [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im]], [[0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] … [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im]; [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] … [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im]; … ; [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] … [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im]; [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] … [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im] [0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im; 0.0 + 0.0im 0.0 + 0.0im 0.0 + 0.0im]]])Plot intensities for a path through $𝐪$-space.
qs = [[0,0,0], [1,0,0]]
path = q_space_path(cryst, qs, 400)
res = intensities_bands(swt, path)
plot_intensities(res; units)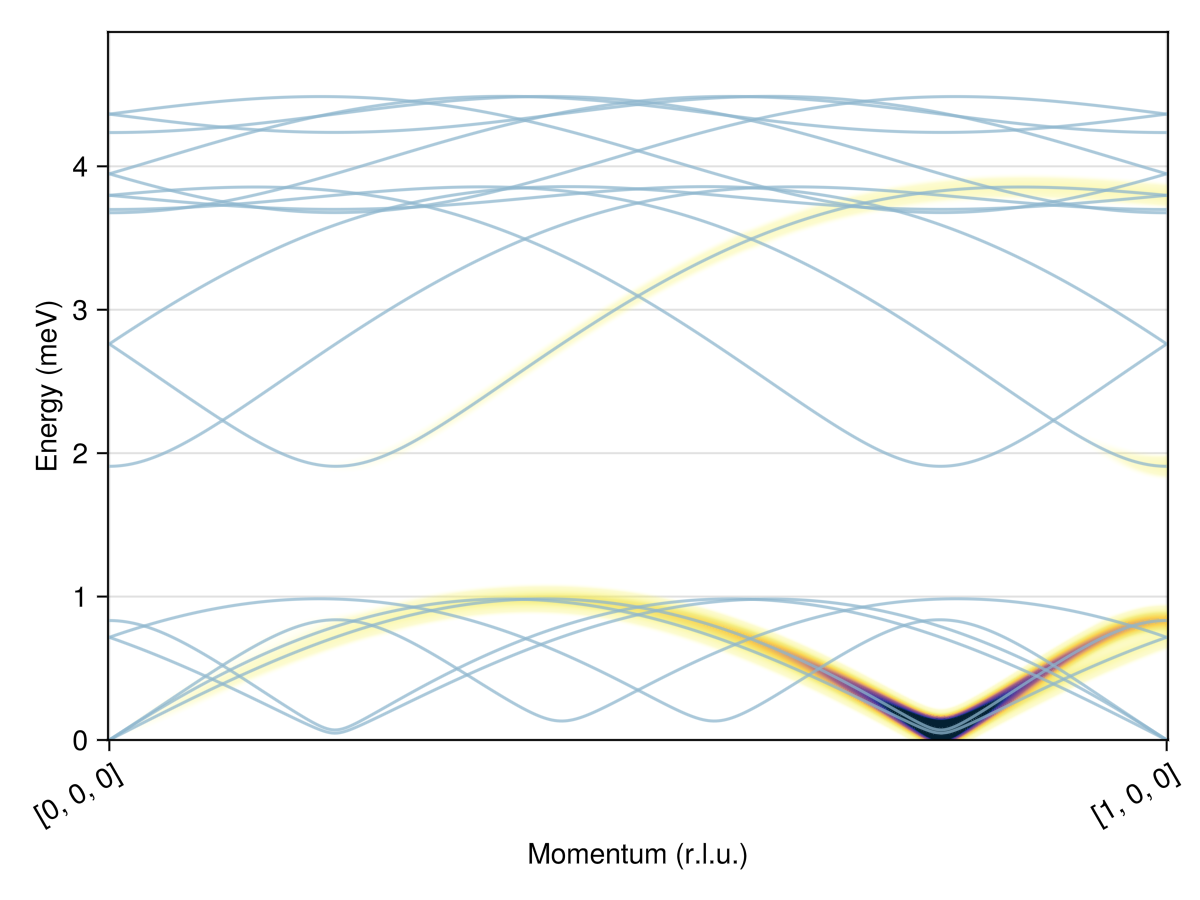
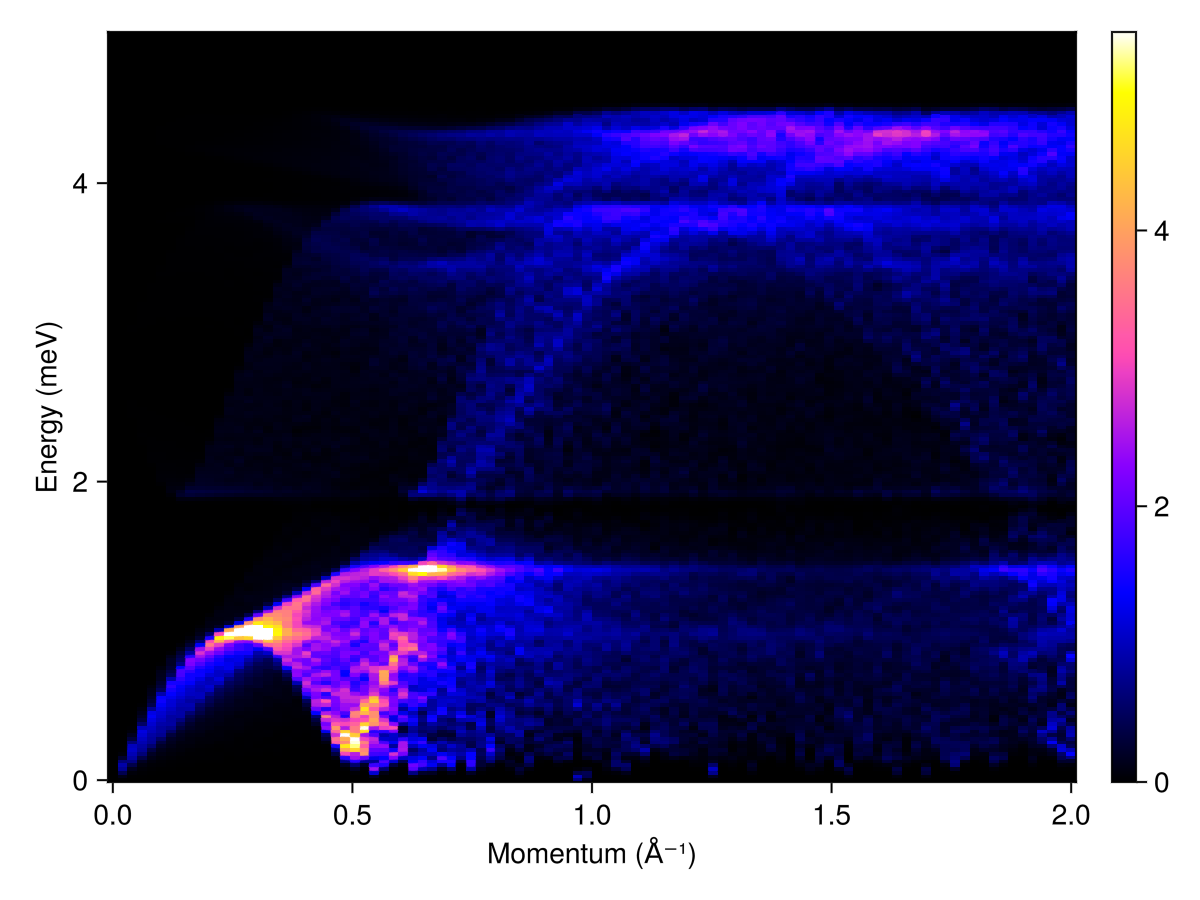
Plot the powder-averaged intensities
radii = range(0, 2, 100) # (1/Å)
energies = range(0, 5, 200)
kernel = gaussian(fwhm=0.05)
res = powder_average(cryst, radii, 400) do qs
intensities(swt, qs; energies, kernel)
end
plot_intensities(res; units)